Plants' oil-desaturating enzymes pair up to channel metabolites
Study raises possibility of linking up enzymes to tailor pathways for increased production of useful plant products
Plant scientists at the U.S. Department of Energy's Brookhaven National Laboratory have found that certain enzymes responsible for desaturating fatty acids, the building blocks of oils, can link up to efficiently pass intermediate products from one enzyme to another. "Engineering these enzyme interactions to channel metabolites along desired metabolic pathways could be a new approach for tailoring plants to produce useful products," said Brookhaven biochemist John Shanklin, lead author on a paper reporting the results in the Journal of Biological Chemistry.
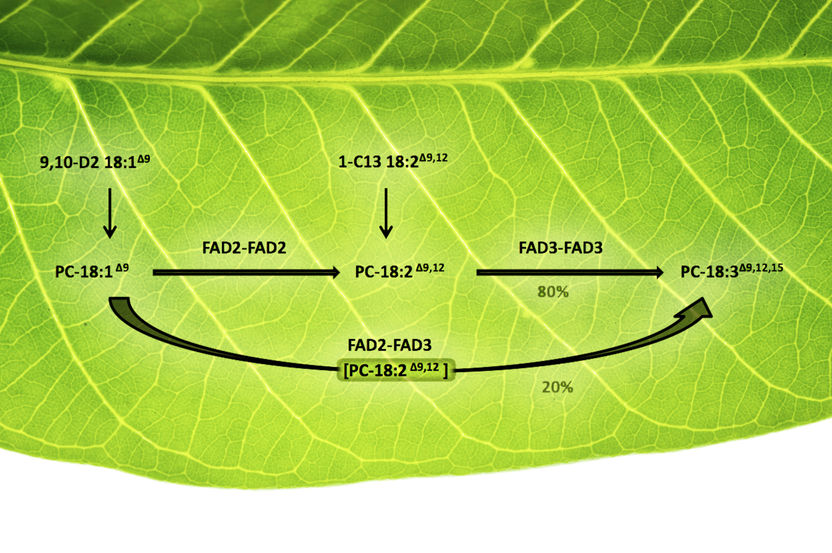
Scientists traced the pathways by which plant oils were desaturated by fatty acid desaturase (FAD) enzymes. Two subsequent desaturating steps could be performed by two different "homodimers"—pairs of identical FADs (FAD2-FAD2 followed by FAD3-FAD3)—or by a heterodimer comprised of FAD2 associated with FAD3. This latter pathway channeled the fatty acid through both desaturation steps without releasing the intermediate compound into the general cell metabolite pool. Engineering such enzyme interactions could offer new ways to direct plant metabolism towards new desired products—for example, polyunsaturated oils or rare fatty acids that could serve as feedstocks for industrial processes.
Brookhaven National Laboratory
Getting plants to accumulate high levels of more healthful polyunsaturated fatty acids, or unusual fatty acids that could be used as raw materials in place of petroleum-derived chemicals in industrial processes, are a few possible outcomes.
Metabolic channeling
The idea would be to take advantage of a process called metabolic channeling, wherein enzymes that act sequentially in a particular metabolic pathway interact with each other so that they are able to pass molecules to each other without them entering the general metabolite pool of the cell. This close arrangement of enzymes also prevents intermediates from entering the metabolic channel.
Previous studies by Shanklin's group had shown that a distinct kind of desaturase enzyme that floats freely in plant plastids—mini plant cell "organs" where many metabolic processes, including photosynthesis, take place—pair up with themselves to form structures called dimers. The group had also studied baker's yeast and determined that its membrane-bound desaturase formed dimers too. But no studies had looked for these kinds of macromolecular arrangements in membrane-bound desaturases, called FADs, in higher plants. The current study used a molecular-genetic approach to explore the organization of membrane desaturases found in the plastids and endoplasmic reticulum (another membrane structure inside cells) of Arabidopsis, a common experimental plant.
Ying Lou, a postdoctoral research fellow working in Shanklin's lab, used several independent methods of bi-molecular complementation—methods that produce a signal if two test proteins come together—to establish which desaturases interact with themselves or others.
The scientists found that all the plant membrane desaturases they examined are capable of forming self-associating dimers in plant cells—pairings of two identical desaturase enzyme molecules. They also found that certain desaturases with different functions could also pair up, but others could not.
"The naturally pairing enzymes turn out to have interesting patterns," Shanklin said. "They are found in the same subcellular locations within the cell, and are involved in subsequent steps of the same metabolic pathway, suggesting a physiological driver for the observed pairings.
"Other pairings between very similar desaturases from different locations that we expected to pair up didn't," he added.
To test the idea that the paired enzymes were working together, the scientists conducted another series of experiments called metabolic flux analysis, drawing on Brookhaven biochemist Jorg Schwender's expertise. This method follows mass-labeled compounds through the various reaction pathways.
"Think of a city map with lots of ways to get from A to B. This method traces how many molecules travel along each route," Schwender said.
The analysis showed that one of the natural enzyme pairings performed two steps of a particular metabolic process without releasing an intermediate product.
"This was clear evidence that these two linked enzymes were working in concert to channel metabolites through this metabolic pathway in an efficient manner in living plant cells," Shanklin said. "Our findings suggest genetic techniques may be used to engineer these kinds of interactions into other desaturase enzymes—including enzymes that don't associate naturally—to push metabolites along desired pathways to produce useful products."