Researchers combine disciplines, computational programs to determine atomic structure
A team from the University of Illinois at Urbana-Champaign and Indiana University combined two techniques to determine the structure of cyanostar, a new abiological molecule that captures unwanted negative ions in solutions.
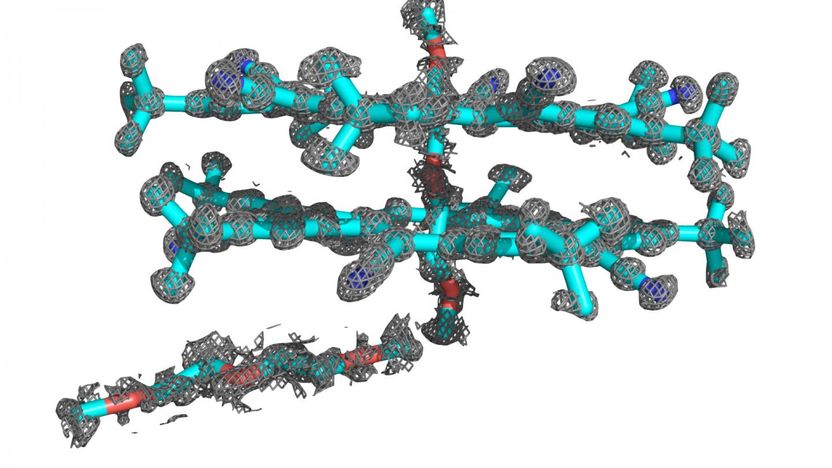
Researchers, led by a team from the Beckman Institute at the University of Illinois, combined the power of two computational programs to determine the atomic structure of the abiological molecule cyanostar. This breakthrough will allow researchers to investigate the structure of more abiological molecules, which are relatively unknown.
University of Illinois
When Semin Lee, a chemist and Beckman Institute postdoctoral fellow at Illinois, first created cyanostar at Indiana University, he knew the chemical properties, but couldn't determine the precise atomical structure. Lee synthesized cyanostar for its unique ability to bind with large, negatively charged ions, which could have applications such as removing perchlorate, a commercially produced contaminate that has been associated with negative effects on human health, from water and soil.
To achieve the highest degree of accuracy in determining the structure of a molecule, scientists have combined methods from both disciplines to determine the complicated structure of cyanostar, a symmetrical, five-sided macrocycle that is easy to synthesize and could also be useful in lithium ion batteries. These results are detailed in the Journal of the American Chemical Society.
"There are two worlds which determine structure - the world of biology, which has big systems but often doesn't generate precise structural calculations," said Klaus Schulten, head of the Theoretical and Computational Biophysics Group (TCBG) at the Beckman Institute for Advanced Science and Technology at Illinois.
"Then there's the chemical world of smaller molecules, which results in super-precise calculations, but often can't account for disorder in the molecule."
To overcome the challenges of the disorder, Lee collaborated with Schulten's group, including Abhishek Singharoy, another Beckman Institute postdoctoral fellow who specializes in a computational simulation method called xMDFF (Molecular Dynamics Flexible Fitting). The method uses x-ray crystallography data to examine a molecule's structure.
The team combined xMDFF with PHENIX, a popular tool in the crystallography community developed by a group at the University of California, Berkeley.
"It's really an excellent combination of the two programs. We wouldn't have gotten the results we did without this computational partnership," Schulten said. "This entire project is really a wonderful example of how today's science is being done. The disciplines joined, the teams joined, and the computational programs joined. When you are open-minded and look to see what others can do, you are really better off."
Topics
Organizations
Other news from the department science

Get the chemical industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.


























































