Figuring out how we get the nitrogen we need
Figuring out how we get the nitrogen we need Caltech chemists image nitrogenase's active site at work
nitrogen is an essential component of all living systems, playing important roles in everything from proteins and nucleic acids to vitamins. It is the most abundant element in Earth's atmosphere and is literally all around us, but in its gaseous state, N2,, it is inert and useless to most organisms. Something has to convert, or "fix," that nitrogen into a metabolically usable form, such as ammonia. Until about 100 years ago when an industrial-scale technique called the Haber-Bosch process was developed, bacteria were almost wholly responsible for all nitrogen fixation on Earth (lightning and volcanoes fix a small amount of nitrogen). Bacteria accomplish this important chemical conversion using an enzyme called nitrogenase.

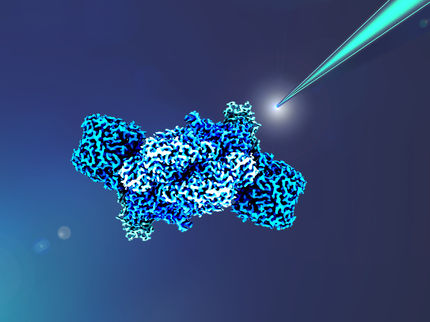
Caltech chemists have taken a crucial step toward unlocking the mystery of how bacteria use an enzyme called nitrogenase to convert nitrogen -- an essential component of all living systems - into a form that living systems can use.
Lance Hayashida / Caltech Marketing and Communications
"For decades, we have been trying to understand how nitrogenase can interact with this inert gas and carry out this transformation," says Doug Rees, Caltech's Roscoe Gilkey Dickinson Professor of Chemistry and an investigator with the Howard Hughes Medical Institute (HHMI). To fix nitrogen in the laboratory, the Haber-Bosch process requires extremely high temperatures and pressures, yet bacteria are able to complete the conversion under physiological conditions. "We'd love to understand how they do this," he says. "It's a great chemical mystery."
But cracking that mystery has proven extremely difficult using standard chemical techniques. We know that the enzyme is made up of two proteins, the molybdenum iron (MoFe-) protein and the iron (Fe-) protein, which are both required for nitrogen fixation. We also know that the MoFe-protein consists of two metal centers and that one of those is the FeMo-cofactor (also known as "the cofactor") at the active site, where the nitrogen binds and the chemical transformation takes place.
In 1992, Rees and his graduate student, Jongsun Kim (PhD '93), were the first to determine the structure of the MoFe-protein using X-ray crystallography.
"I think that there was a feeling that once you solved the structure, you'd understand how it worked," Rees says. "What we can say 22 years later is that was certainly not the case."
The dream would be to have atmospheric nitrogen bind to the FeMo-cofactor and to stop time so that chemists could sneak a peak at the chemical structure of the protein at that intermediate point. Since it is not possible to freeze time and because the reaction proceeds too quickly to study by standard crystallographic methods, researchers have come up with an alternative. Chemists have been trying to get carbon monoxide, an inhibitor that halts the enzyme's activity but also closely mimics the structure and electronic makeup of N2, to bind to the cofactor and to then crystallize the product relatively quickly so that the structure can be analyzed using X-ray crystallography.
Unfortunately, the cofactor has stubbornly refused to cooperate. "We've demonstrated more times than we'd like that the form of this protein as isolated doesn't bind substrates," explains Rees. "Usually if you want to know how something binds to a protein, you just add it to your protein and study the crystal structure with X-ray crystallography. But we just couldn't get anything bound to this cofactor."
But in order for the cofactor to exist in a form that would bind to a substrate or an inhibitor, several other conditions must be met—for example, the Fe-protein has to be there. In addition, ATP—a molecule that provides energy for many life processes—must be present, along with yet another enzyme system that regenerates the ATP consumed in the reaction and a source of electrons. So although the aim in crystallography is typically to isolate a purified protein, the chemists had to muddy their samples by adding all these other needed components.
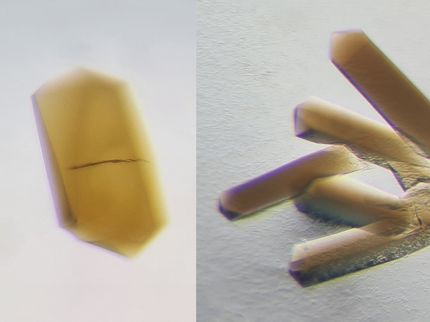
After joining Rees's group as a postdoctoral scholar in 2012, Thomas Spatzal spent months working on this problem, tweaking the method he used for trying to get the carbon monoxide to bind to the cofactor and for crystallizing the product. He adjusted parameters such as the protein concentrations, the temperature under which the samples were prepared, and the amount of time he allowed for the crystals to form. Every week, he sent a new set of crystals, frozen with liquid nitrogen, to be analyzed on an X-ray beamline at the Stanford Synchrotron Radiation Lightsource (SSRL) constructed as part of Caltech's Molecular Observatory with support from the Gordon and Betty Moore Foundation. And every week he worked up the data that came back and looked to see if any of the carbon monoxide bound to the active site.
"People have been seeing the resting state of the active site, where nothing was bound, for years," Spatzal says. "It's always the same thing. It never looks any different."
But on a recent Friday morning, Spatzal processed the latest batch of data, and lo and behold, he finally saw what he had been looking for.
"There was a moment where I looked at it and said, 'Hold on. Something looks different there,'" says Spatzal. "I wondered, 'Am I crazy?' You just don't expect it at first."
What he saw was a first—a crystal structure revealing carbon monoxide bound to the FeMo-cofactor. Spatzal, Rees, and their colleagues describe that structure and their methodology in the September 26 issue of the journal Science.
Spatzal figured out a way to optimize the crystallization process by using tiny crystal seeds to accelerate the rate of crystal growth and conducting all manipulations in the presence of carbon monoxide, allowing him to grow nice crystals of the MoFe-protein and then to see where the carbon monoxide was bound to the cofactor.
What he found was surprising. The carbon monoxide took the place of one of the sulfur atoms in the cofactor's original structure, bridging two of its iron atoms. Many people had expected that the carbon monoxide would bind differently, so that it would stick out, adding extra density to the structure. But because it displaced the sulfur, the cofactor only took on a slightly different arrangement of atoms.
In addition, Spatzal showed that when the carbon monoxide is removed, the sulfur can reattach, reactivating the cofactor so that it can once again fix nitrogen.
"As astonishing as this structure was—that the carbon monoxide replaced the sulfur—I think it's even more astonishing that Thomas was able to establish that the cofactor could be reactivated," Rees says. "I don't think anyone had imagined that you would get this sort of rearrangement of the cofactor as part of the interaction."
"You could imagine that if you put an inhibitor on a system, it could damage the metal center and inactivate the protein so that it would no longer do its job. The fact that we can get it back into an active state means that it's not permanently damaged, and that has physiological meaning in terms of how nitrogen fixation occurs in nature," says Spatzal.
Original publication
Ligand binding to the FeMo-cofactor: Structures of CO-bound and reactivated nitrogenase ,"