'Semi-synthetic' bacteria churn out unnatural proteins
Advertisement
Synthetic biologists seek to create new life with forms and functions not seen in nature. Although scientists are a long way from making a completely artificial life form, they have made semi-synthetic organisms that have an expanded genetic code, allowing them to produce never-before-seen proteins. Now, researchers reporting in Journal of the American Chemical Society have optimized a semi-synthetic bacteria to efficiently produce proteins containing unnatural amino acids.
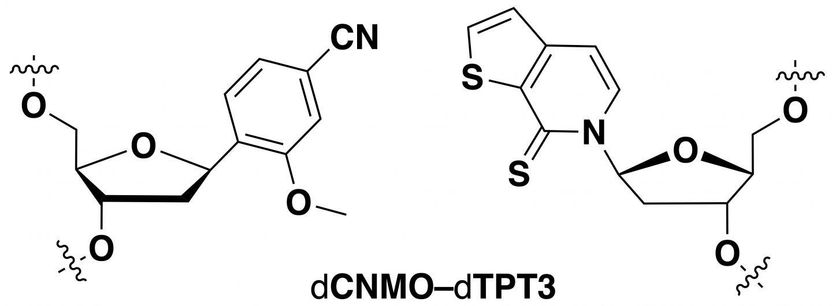
Researchers identified this unnatural base pair as being optimal for information storage in a semi-synthetic organism.
Adapted from Journal of the American Chemical Society 2019, DOI: 10.1021/jacs.9b02075
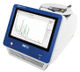
All of Earth's natural life forms store information using a four-letter genetic code consisting of the nucleotides deoxyadenosine (dA), deoxyguanosine (dG), deoxycytidine (dC), and deoxythymidine (dT). Within the DNA double helix, dA pairs with dT, and dG with dC, to form the "rungs" of the DNA ladder. Recently, researchers have made synthetic nucleotides that can pair up with each other. When they placed these unnatural nucleotides into genes, bacteria could replicate the DNA and convert the sequences into RNA and then proteins that contained unconventional amino acids. However, bacteria often cannot use these synthetic sequences as efficiently as the natural ones. Therefore, Lingjun Li, Floyd Romesberg and colleagues wanted to optimize the unnatural base pairs to improve protein production.
The researchers tested different combinations of unnatural base pairs in E. coli and observed which ones were replicated most efficiently and produced the highest levels of a protein. Some of the synthetic base pairs had been tested before, whereas others were new variations. The team then used these optimized base pairs to demonstrate, for the first time, a semi-synthetic organism that could make a protein containing multiple unnatural amino acids.
Original publication
Original publication
Aaron W. Feldman et al.; "Optimization of Replication, Transcription, and Translation in a Semi-Synthetic Organism"; J. Am. Chem. Soc.; 2019
Organizations
Other news from the department science

Get the chemical industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.